I am very pleased to announce that myself, along with Aki Watanabe and Marc Jones will be leading a Symposium and associated workshop on Geometric Morphometrics in Paleontology at the Annual Meeting for the Society of Vertebrate Paleontology in Dallas Texas October 2015.
Podium Symposium: The Shape of Things to Come: Geometric Morphometrics in Vertebrate Paleontology. Details are here. The oral presentations (invited speakers only) will take place in one of the morning sessions, with 16 speakers, which we believe will provide you with a broad range of theory and applications of GMM in vert paleo.
The Workshop: Geomorph: R Package for the Collection and Analysis of Geometric Morphometric Data, is scheduled for Tuesday, October 13, from 9:30 am – 4:30 pm, at the Hyatt Regency Dallas. Registration for the workshop costs $40 and is open to all SVP 2015 participants. Places are limited to 20 participants.
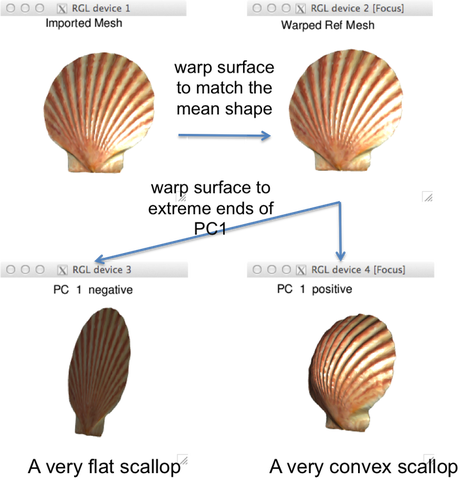
The purpose and intended outcome of the workshop: Geometric morphometrics (GMM) has become an increasingly popular method for quantifying and analyzing the morphology of specimens in vertebrate paleontology. The integration of GMM data with multivariate statistics and phylogenetic comparative methods makes it a particularly powerful and versatile technique for tackling biological questions related to shape. Recently, the proliferation of available software for collecting, analyzing, and visualizing shape data, especially in 3-D, has driven this growing adoption of GMM. The primary objective of the workshop is to equip the participants with the competency to collect and analyze GMM data in the freely and openly available “geomorph” R package. Topics covered will be largely practical with some theoretical components, including (1) digitization of specimens; (2) superimposition techniques; (3) multivariate statistical methods; and (4) comparative phylogenetic methods. The workshop expects the participants to bring their own laptops and have some proficiency with R statistical software. Although example data will be provided, participants are encouraged to bring their own data sets. From this workshop, the participants will acquire their own GMM data set, in addition to the skill and resources to execute analyses on their shape data.
Hope to see you there!
Emma
Podium Symposium: The Shape of Things to Come: Geometric Morphometrics in Vertebrate Paleontology. Details are here. The oral presentations (invited speakers only) will take place in one of the morning sessions, with 16 speakers, which we believe will provide you with a broad range of theory and applications of GMM in vert paleo.
The Workshop: Geomorph: R Package for the Collection and Analysis of Geometric Morphometric Data, is scheduled for Tuesday, October 13, from 9:30 am – 4:30 pm, at the Hyatt Regency Dallas. Registration for the workshop costs $40 and is open to all SVP 2015 participants. Places are limited to 20 participants.
The purpose and intended outcome of the workshop: Geometric morphometrics (GMM) has become an increasingly popular method for quantifying and analyzing the morphology of specimens in vertebrate paleontology. The integration of GMM data with multivariate statistics and phylogenetic comparative methods makes it a particularly powerful and versatile technique for tackling biological questions related to shape. Recently, the proliferation of available software for collecting, analyzing, and visualizing shape data, especially in 3-D, has driven this growing adoption of GMM. The primary objective of the workshop is to equip the participants with the competency to collect and analyze GMM data in the freely and openly available “geomorph” R package. Topics covered will be largely practical with some theoretical components, including (1) digitization of specimens; (2) superimposition techniques; (3) multivariate statistical methods; and (4) comparative phylogenetic methods. The workshop expects the participants to bring their own laptops and have some proficiency with R statistical software. Although example data will be provided, participants are encouraged to bring their own data sets. From this workshop, the participants will acquire their own GMM data set, in addition to the skill and resources to execute analyses on their shape data.
Hope to see you there!
Emma






 RSS Feed
RSS Feed
